The availability of large amounts of data has revolutionized research in the life sciences in the past few years, offering a wide range of opportunities for knowledge gain and future applications. By combining the disciplines of mathematics, computer science, medicine and life sciences, bioinformatics has made it possible to store, categorize, analyze, evaluate and visualize biological data and to simulate biochemical processes.
Applied bioinformatics and artificial intelligence
Getting the most out of big data for biomedical translation
Application areas at Fraunhofer ITEM
At Fraunhofer ITEM, researchers develop methods and possibilities for the preparation, analysis and visualization of biomedical data, as well as data models and data analysis pipelines. The focus of our research is on the mapping of cellular and regulatory processes and their translation into applications for humans. Bioinformatics methods are used, for example, for personalized tumor therapy to develop optimized testing strategies and for research on RNAs as diagnostic biomarkers and therapeutic targets. For personalized therapies or for patient stratification, the knowledge gained from big data is key to identifying adequate treatment strategies. Stratification also plays a major role for hazard and risk assessment of chemicals, nanomaterials, and environmental exposure, as the sensitivity to noxious agents differs between subpopulations.
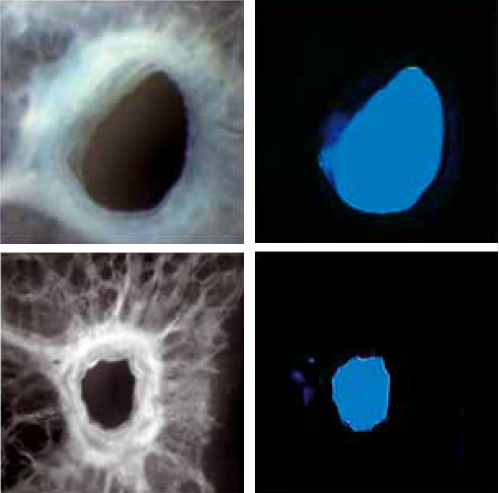
In addition, the Fraunhofer researchers are using bioinformatics and artificial intelligence to advance towards intelligent image data analysis and are further developing this technology, so as to optimize the analysis of histological images and support clinical processes.
 Fraunhofer Institute for Toxicology and Experimental Medicine
Fraunhofer Institute for Toxicology and Experimental Medicine